Published online: October 2022
Synthesis by: Natalie Forsdick, Manaaki Whenua - Landcare Research (forsdickn @ landcareresearch.co.nz, Twitter: @NatForsdick); Aisling Rayne, School of Biological Sciences, University of Canterbury; Molly Magid, School of Biological Biological Sciences, University of Canterbury; Tammy Steeves, School of Biological Sciences, University of Canterbury
To achieve the vision of thriving biodiversity outlined in Te Mana o te Taiao, Aotearoa New Zealand’s National Biodiversity Strategy, we need to bring together diverse knowledge systems, expertise, and disciplines. Among these is conservation genetics/genomics, which uses characteristics of DNA to inform conservation management. Some characteristics common across small, threatened populations include low genetic diversity (which can restrict a species’ potential to adapt to future changes, like new diseases or warming waters), and inbreeding depression (where breeding between close relatives reduces survival or reproductive success). Both can limit species recovery.
Genetic marker sets typically comprise small numbers of short variable regions of DNA such as microsatellites. Despite only capturing a fraction of variation in the genome (the entire set of genetic material present in an organism), genetic markers have been widely used to examine characteristics like low genetic diversity and inbreeding. Technology has improved rapidly in the last two decades, enabling the generation of many thousands of high-resolution genomic markers (such as single nucleotide polymorphisms, or SNPs) with broad conservation applications.
With a growing conservation genetic/genomic toolbox, it can be challenging to choose the best tool for a given application. Options may also be limited by available time, resources/funding, and expertise. Tools currently in use in Aotearoa include pedigrees, genetic and genomic markers, whole genomes, environmental DNA (eDNA), ancient DNA (aDNA), transcriptomes, and microbiomes (see Figure). Each presents unique opportunities, challenges, and trade-offs for conservation research. In this dynamic landscape, it is important for conservation practitioners and researchers to share their expertise when selecting conservation genetics/genomics tools that are fit for purpose. For example, while genetic markers may be sufficient for determining parentage in many threatened species such as kakī/black stilts, genomic markers may be required to investigate characteristics of genetically depauperate species, such as kēkēwai/wai kōura/freshwater crayfish. Costs associated with genomic tools are continuing to decline, and ongoing improvements in these technologies are producing more comprehensive and accurate data, making them a competitive alternative to traditional genetic markers.
What lies ahead for the conservation toolbox? Existing tools are being applied in new ways, such as the combination of ancient and environmental DNA techniques to infer the past distribution of species with relevance to restoration ecology. A range of new tools are also in development. For example, work is currently underway to explore the impacts of structural variants on genetic diversity (where structural variants are rearrangements of DNA in the genome that can play an important role in determining an organism’s form and function). Other tools like genome editing or biobanking (cryopreservation) face substantial technical, legislative, and social challenges before they may be applied for conservation in Aotearoa.
For taonga (treasured) species in Aotearoa, any work involving conservation genetics/genomic tools should begin from a foundation of trusted relationships with mana whenua. Centring community needs, aspirations, and expertise can produce diverse benefits for all involved, including improved research and conservation outcomes. There are a growing number of national and international resources available to guide such partnerships, including frameworks for iterative engagement, and data management practices that centre Indigenous data sovereignty. It is through these kinds of ethical and collaborative approaches that we will be best placed to achieve the aspirational vision of Te Mana o te Taiao.
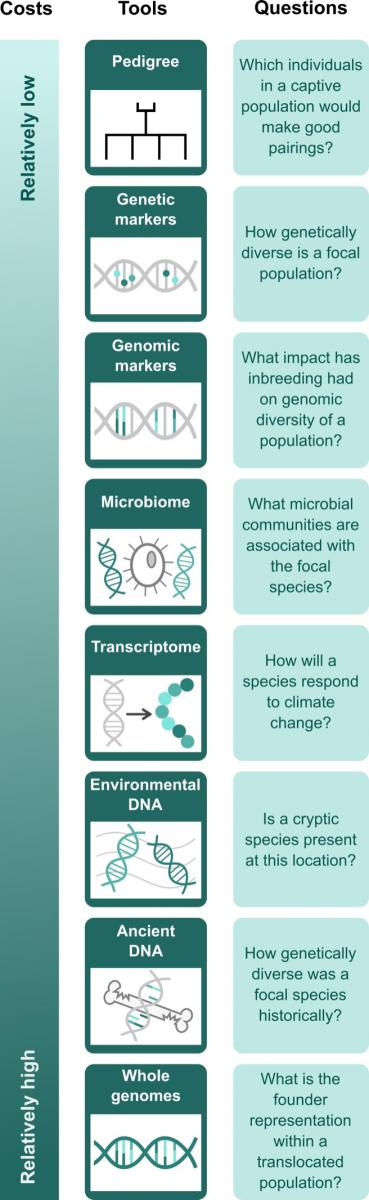
A broad overview of the types of genetics/genomics tools applied to conservation research in Aotearoa New Zealand, including the relative costs (which not only include funding, but also time, invasiveness of sampling, laboratory equipment/resources, computer and data storage resources, and expertise), as reviewed by Forsdick et al. (2022). Pedigrees are family trees tracking the relationships between individuals, and are most commonly used to support conservation breeding programmes in captive management settings. Genetic data are representative of a subset of the genome comprising tens of markers, while genomic data comprise hundreds to millions of markers. Whole genomes can be used for population- or species-level comparisons to unravel phylogenetic relationships and evolutionary pathways. The microbiome refers to the community of microorganisms (fungi, bacteria, viruses) that live on or in a host species. By using transcriptomes we can understand gene expression underpinning the form and function of organisms. DNA present in the environment can be used to characterise current biodiversity, while DNA extracted from historical specimens and fossils can provide a window into the past to inform future conservation and restoration efforts
Relevant literature:
Collier-Robinson L, Rayne A, Rupene M, Thoms C & Steeves T (2019) Embedding Indigenous principles in genomic research of culturally significant species: a conservation genomics case study. New Zealand Journal of Ecology 43. https://doi.org/10.20417/nzjecol.43.36
Forsdick NJ, Adams CIM, Alexander A, Clark AC, Collier-Anderson L, Cubrinovska I, Croll Dowgray M, Dowle EJ, Duntsch L, Galla SJ et al. (2022) Current applications and future promise of genetic/genomic data for conservation in an Aotearoa New Zealand context. Science for Conservation 367. Department of Conservation, New Zealand. https://www.doc.govt.nz/globalassets/documents/science-and-technical/sfc337entire.pdf
Hudson M, Thompson A, Wilcox P, Mika J, Battershill C, Stott M, Tūrongo Brooks R & Warbrick L (2021) Te Nohonga Kaitiaki Guidelines for genomic research on taonga species. https://www.genomics-aotearoa.org.nz/sites/default/files/2022-02/Te%20Nohonga%20Kaitiaki%20Guidelines%20for%20genomic%20research%20on%20taonga%20species%20%28with%20background%29.pdf
Liggins L, Arranz V, Braid HE, Carmelet-Rescan D, Elleouet J, Egorova E, Gemmell MR, Hills SFK, Holland LP, Koot EM et al. (2022) The future of molecular ecology in Aotearoa New Zealand: an early career perspective. Journal of the Royal Society of New Zealand 0, 1–24. https://doi.org/10.1080/03036758.2022.2097709
Mc Cartney AM, Anderson J, Liggins L, Hudson ML, Anderson MZ, Te Aika B, Geary J, Cook-Deegan R, Patel HR, Phillippy AM (2022) Balancing openness with Indigenous data sovereignty: An opportunity to leave no one behind in the journey to sequence all of life. Proceedings of the National Academy of Sciences 119, e2115860119. https://doi.org/10.1073/pnas.2115860119
Te Mana o Te Taiao - Aotearoa New Zealand Biodiversity Strategy 2020. (2020) Department of Conservation, New Zealand. https://www.doc.govt.nz/nature/biodiversity/aotearoa-new-zealand-biodiversity-strategy/te-mana-o-te-taiao-summary/
